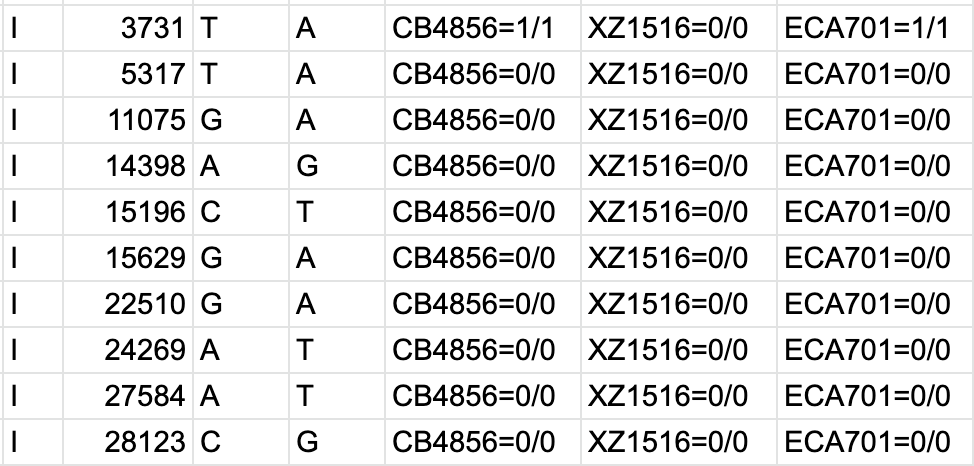
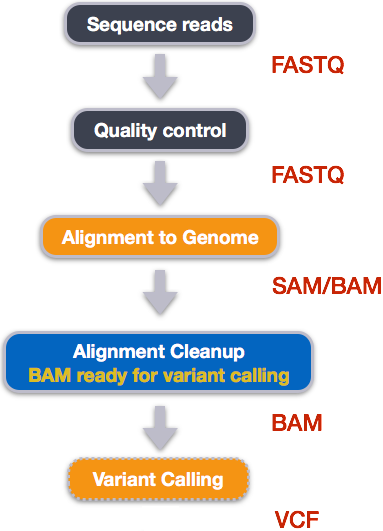
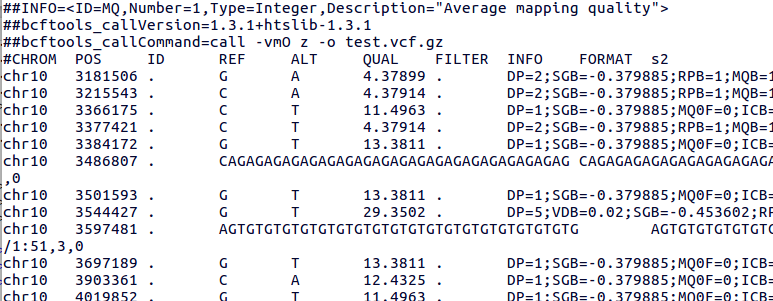
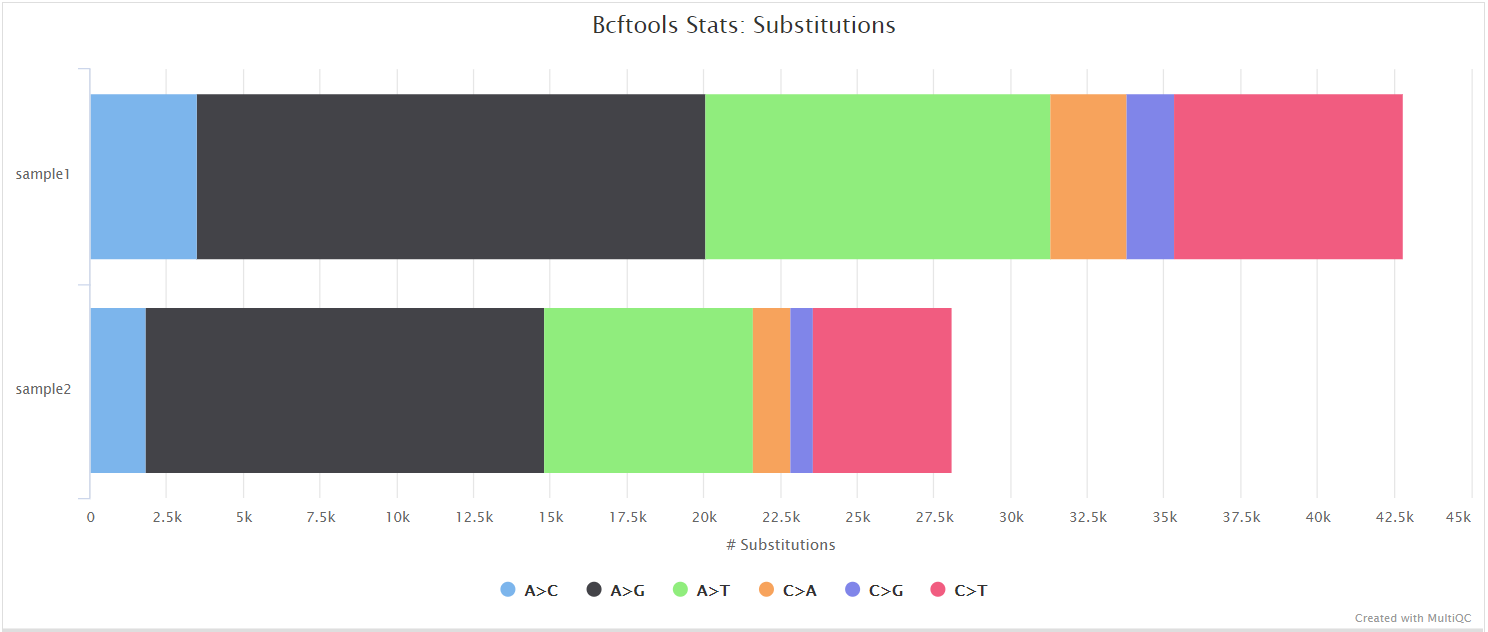
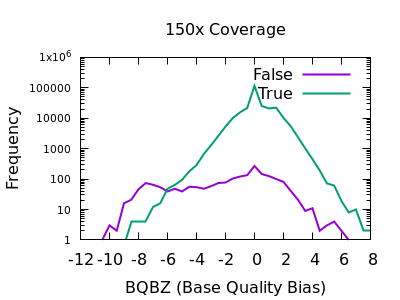
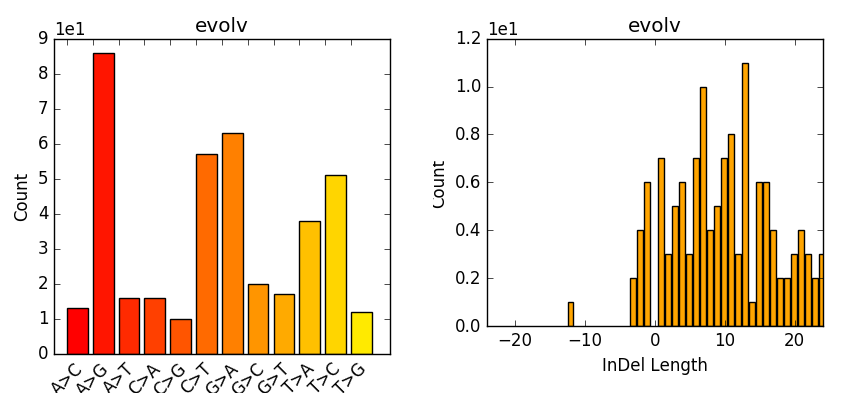
bcftools view | bcftools tutorial on how to count the number of snps and indels in a vcf file - YouTube

Comparison of error rates of BCFtools/RoH and other existing methods as... | Download Scientific Diagram

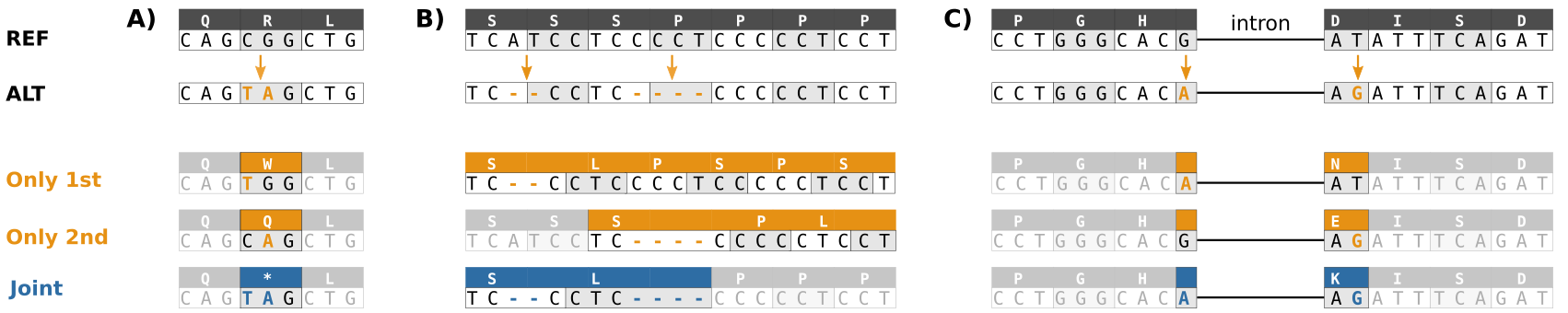
bcftools mpileup - Difference between IDV and FORMAT/AD* fields · Issue #912 · samtools/bcftools · GitHub

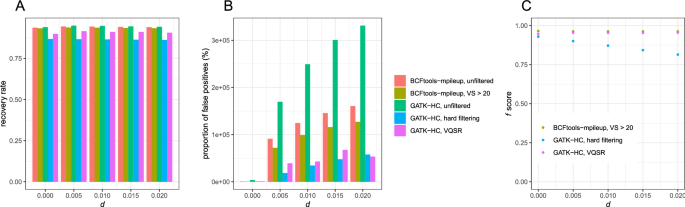
Comparison of selectivity and sensitivity of three SNP-calling tools... | Download Scientific Diagram